Our research
Comparing genomes
We work on fast methods for genome comparison. These comparisons are
based on the lengths of maximal matches, which are matches that cannot
be extended without losing the match. For example, if we compare the
two sequence S_1=AAGGCCCTCT and S_2=AAGGGCCCTCT, the maximal match
starting at the first position in S_1 with respect to S_2 is AAGG
and has length 4.
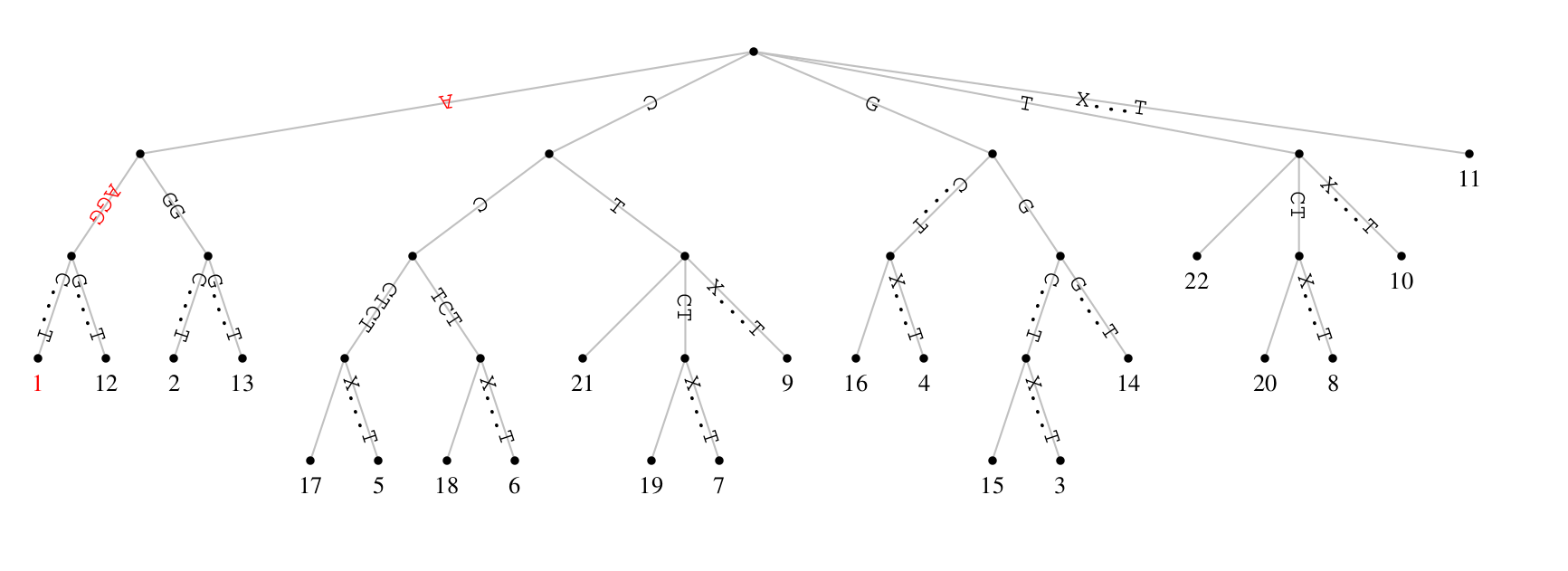
Maximal matches can be looked up quickly from their suffix tree. The
figure above shows the suffix tree for our concatenated example
sequences, S_1XS_2. The path leading from the root to a leaf spells
out the suffix starting at the leaf label. For example, the path
leading to leaf 1 on the left is labeled with the complete input,
S_1XS_2. To look up the maximal repeat starting at a some position,
visit the corresponding leaf and look up the path label of its
parent. In our example, the leaf position is 1 and the parent’s leaf
label the AAGG we also found by directly comparing S_1 and S_2.
The ease with which one can locate maximal matches using suffix trees becomes interesting when we realize that all maximal matches end in a mismatch. Mismatches, or single nucleotide polymorphisms, SNPs, underlie all of comparative genomics. So in order to identify SNPs, we just have to count homologous, as opposed to random, matches.
To identify homologous matches, we derived the null
distribution of maximal match
lengths. Based on this and fast suffix tree construction, we
developed the program
phylonium, for fast and
accurate estimation of pairwise evolutionary distances between
hundreds of bacterial genomes.
We have used a similar approach in our program
fur, for finding unique genomic
regions from whole bacterial genomes. The program fur compares a
sample of target genomes to their closest evolutionary neighbors and
picks the regions present in all targets that are absent from the
neighbors. These regions are good candidates for diagnostic markers.
Looking up genomes
In our work on genome comparison, we rely on efficient methods for
looking up suitable genomes. For this purpose we have developed the
package Neighbors. Given a
target taxon, Neighbors looks up the genomes of the target and
neighbor taxa using the NCBI
taxonomy. However, these
taxonomic targets and neighbors are not necessarily identical to the
desired phylogenetic targets and neighbors. So we calculate the
phylogeny of the taxonomic targets and neighbors using phylonium,
and then use tools from the Neighbors package to sort them into
phylogenetic targets and neighbors.